The scientific data quagmire – siloed, fragmented data in hundreds of proprietary formats, manually retrieved from instruments, edited in spreadsheets – wastes enormous time. Worse, that time is stolen from some of the industry’s most important players: the scientists, data scientists, data engineers, informaticians and analysts who might otherwise use their valuable skills to generate fresh knowledge and improve processes to better discover, develop and deliver new life-saving therapeutics.
Worse yet, even as data volumes continue to grow at astronomical rates, the data quagmire prevents biopharma organizations from effectively applying transformative technologies — like automation, analytics, and AI/ML (Artificial Intelligence and Machine Learning). Technologies that can help them keep up, stay ahead, and generate new and valuable insights. Technologies that unblock new and promising avenues of research, like genome studies, whose vast search spaces can only be effectively explored with machine assistance.
The problem, as CSO Mike Tarselli made clear in a recent interview published by AZO Life Sciences, is that analytics software can only consume what he calls "arranged data" – data in a standardized format, structured, normalized, and contextualized according to well-understood, Big Data rules.
What “Lab of the Future” Would You Build?
Those envisioning, leading, and implementing the next generation of biopharma laboratory evolution are asking themselves how to solve this problem. How do you make scientific data findable, accessible, interoperable, and reusable (FAIR), so that science-accelerators like AI/ML can be put to work in earnest? They're also asking the follow-on question: If we could make the data quagmire go away, what would we build?
Manual No More: Automating the Scientific Data Lifecycle, a new whitepaper from TetraScience CTO Spin Wang and CSO Mike Tarselli, outlines a practical solution to the data quagmire problem. Their proposed method applies well-understood automation and data transformation technology to speed up iterative experimental workflows, increase predictability, eliminate manual data handling, and save scientists’ time. They then expand the strategy – showing how analytics and AI/ML can be inserted into the loop to provide deeper and more complete insights to the organization.
Automating the DMTA Loop with a Data-centric Scientific Platform
The white paper opens with this simple question – originally asked of TetraScience customers, partners, and advisors: "If you built a lab from scratch to orchestrate the free flow of information and data across your laboratory instruments and informatics applications to perform data analysis … what would you build?"
Those asked responded with a tight wish-list focused on automating parts of the so-called Design/Make/Test/Analyze loop (Figure 1).
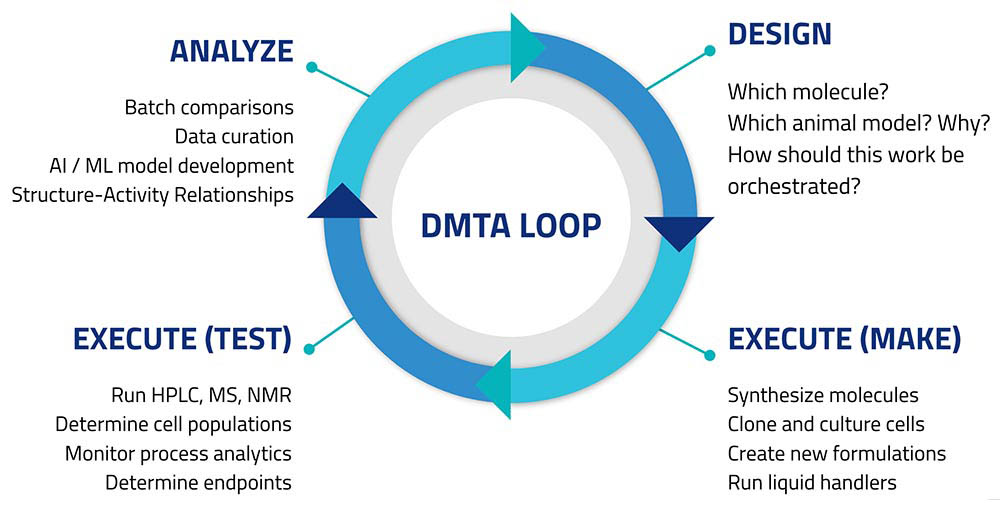
Customers sought to create connectivity among lab informatics systems like ELNs and LIMS, control software for automated instruments (e.g., HPLC, chromatography data systems), robotics, and standalone instruments like balances and pH meters. This would let scientists design experiments in their preferred digital workbench applications, then export these designs to automate experimental runs. Results of experiments, and measurements made on non-networked instruments would all be returned to the informatics platforms; and become accessible from centralized storage.
Customers had requirements for the data too. They wanted raw data extracted, transformed and harmonized into a vendor-neutral format, and stored in an “arranged” way: prepared and formatted to simplify modeling and advanced analytics, REST API-accessible, able to flow among and be used to update multiple systems (e.g., sample management, inventory, etc.), and perhaps already integrated with popular analytics platforms like SpotFire or Tableau, making custom analytics fast and simple.
As the whitepaper makes clear, this requires a scientific data cloud: a data-centric platform designed to extract, transform, and harmonize data, store it, make it accessible, and deliver it to targets reliably, serving the requirements of complex workflows. Part of the benefit of such a system is that it can be used to break scientific data out of "walled garden" silos (e.g. instruments and their control software, which may integrate with popular ELNs, but are seldom easily integrated with other software), separate the data from instrument command/control messaging, and ultimately make the data FAIR, compliant, harmonized, liquid, and actionable.
Two Models for Scientific Data Automation
The whitepaper follows this wish-list to specify two, similar models for basic laboratory automation. In the first model – simpler – scientists, perhaps working with visualization and/or analytics tools, make decisions to guide each Design/Make/Test/Analyze iteration. In the second, software accelerates decision-making. In some scenarios, a scientist may interact with a decision-tree to choose among alternatives suggested by analytics or AI/ML. In others, AI/ML or heuristic software takes over and runs the DMTA loop to a stopping-point.
The models differ, but can be taken to represent successive steps in a lab's progress towards reduced scientist labor, and greater speed and efficiency. The first step makes maximal use of scientist participation. In the second, the scientist (or a number of scientists) serves as the model (for decision-trees and heuristics) or as the de facto trainer of machine-learning models – in effect, porting essential, but low-value work into software to free up staff time for higher-value work.
Moving from Scientific Data Quagmire to the Future of Laboratory Automation
Reimagining scientific data management leads to a new world where FAIR data flows freely between instruments, informatics and analytics systems to accelerate scientific innovation across biopharma discovery, development and delivery. Learning how this works in greater detail can be a first step towards helping your organization navigate out of the data quagmire and into the fast-moving future of laboratory automation.

